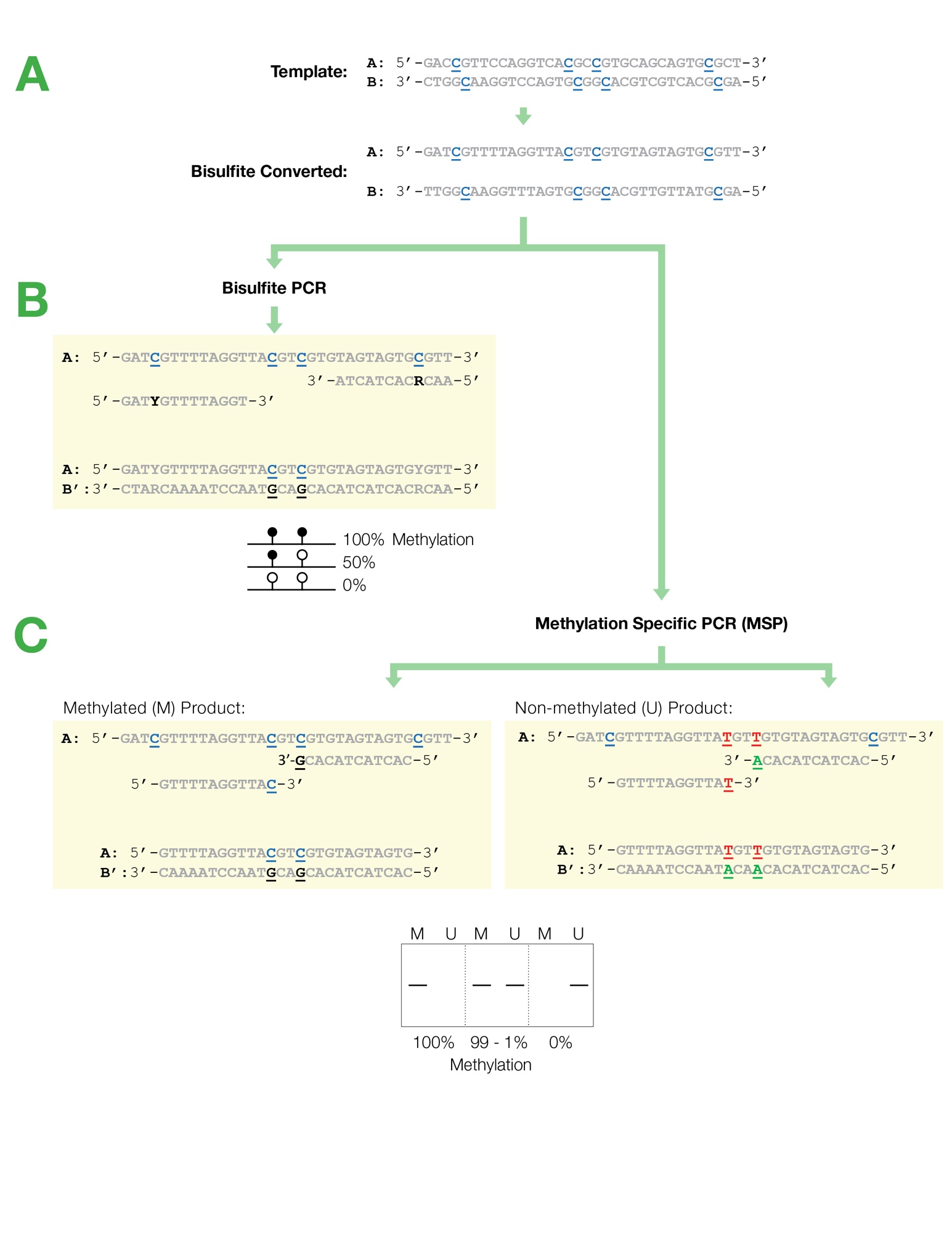
Designing primers for methylation PCRs. B Primers for Bisulfite PCR are designed for subsequent sequencing and analysis of cytosines within the amplicon. Bisulfite sequencing primer design.
Bisulfite Sequencing Primer Design, Unlike normal PCR bisulfite PCR primers need to be long usually between 26-30 bases and the amplicon size. CpG sites within the primers should be avoided or located at the 5-end with a mixed base at the cytosine position Y CT R GA. Successful bisulfite primer design is critical to unbiased region-specific DNA methylation analysis. Multiple options for amplicon length and sequence location within the region of.
 Hints For Optimizing Bisulfite Primer Design Zymo Guide From epigenie.com
Hints For Optimizing Bisulfite Primer Design Zymo Guide From epigenie.com
Pick primers for bisulfite sequencing PCR or restriction PCR. Bisulfite Primer Seeker is a free program that has been optimized to design primers for CGrich sequences where others usually fail. Bisulfite specific primers can be used as one part of a Sanger sequencing or targeted NGS workflow. Bisulfite Primer Design Application.
How to cite MethPrimer.
Read another article:
Bisulfite Primer Seeker is a free program that has been optimized to design primers for CGrich sequences where others usually fail. To design primers for bisulfite-modified DNA users are required to either copy and paste their unmodified sequences into the text area or upload a multi-FASTA file into PS. Ad Design the perfect primers for your PCR CE sequencing and cloning. Existing primer design programs developed for standard PCR cannot handle primer design for bisulfite-conversion-based PCRs due to changes in DNA sequence context caused. OligoPerfect allows design up to 50 primers optimized for your reaction conditions.
 Source: nature.com
Source: nature.com
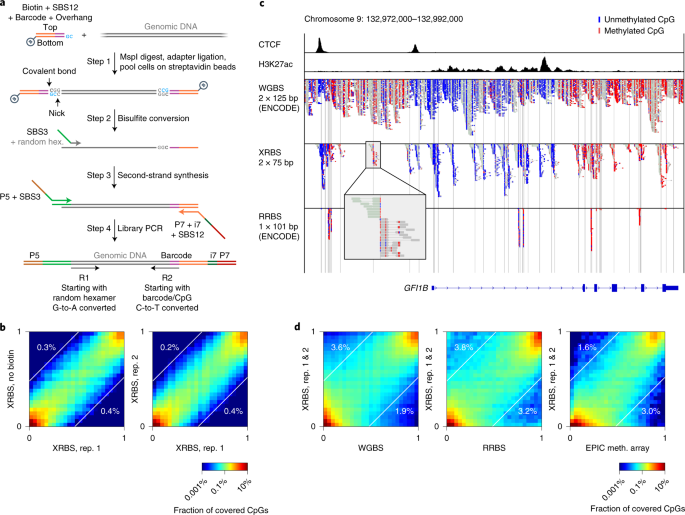
Multiple options for amplicon length and sequence location within the region of. Multiple options for amplicon length and sequence location within the region of. It may be used for both bisulfite converted as well as for original not modified sequences. CpG sites within the primers should be avoided or located at the 5-end with a mixed base at the cytosine position Y CT R GA. Extended Representation Bisulfite Sequencing Of Gene Regulatory Elements In Multiplexed Samples And Single Cells Nature Biotechnology.
 Source: pinterest.com
Source: pinterest.com
The sequences obtained from methylated gDNA and unmethylated gDNA are fundamentally different after bisulfite conversion. The software is able to design primers for the PCR amplification of both bisulfite-treated and untreated sequences. Features of the Bisulfite Primer Seeker Program. Capable of designing primers in CG-rich regions where others usually fail. Benefits And Drawbacks Associated With Medical Utilizing Mammalian Expression Methods Expressions Recombinant Dna Medical.
 Source: nature.com
Source: nature.com
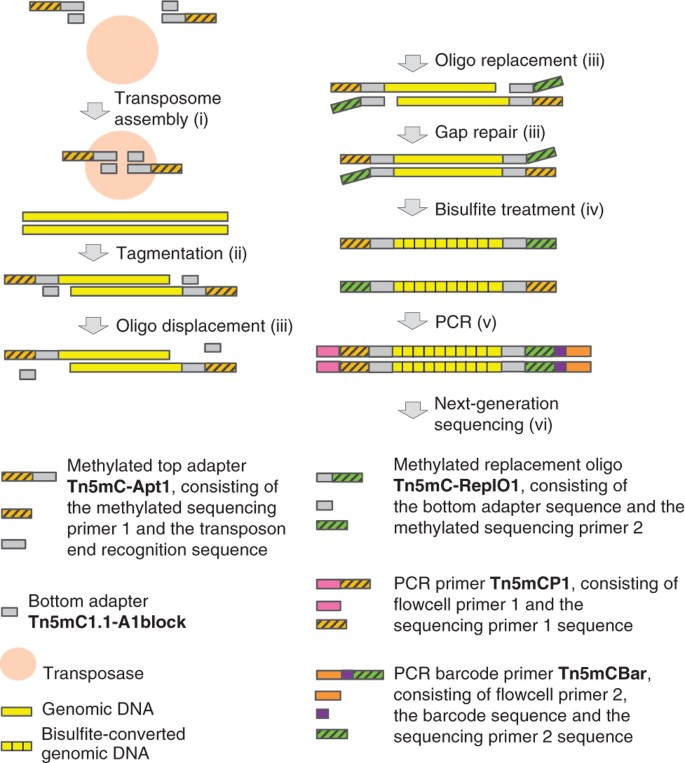
Convert C to T before pasting. Currently it can design primers for two types of bisulfite PCR. You dont need to modify your sequence eg. Li LC and Dahiya R. Tagmentation Based Whole Genome Bisulfite Sequencing Nature Protocols.
 Source: ro.pinterest.com
Source: ro.pinterest.com
Designing Primers While good primer design is critical for successful PCR in any analysis designing primers for methylated DNA has many additional confounding factors to consider. Unlike normal PCR bisulfite PCR primers need to be long usually between 26-30 bases and the amplicon size. BiSearch is a primer-design algorithm for DNA sequences. Currently it can design primers for two types of bisulfite PCR. Pin On Bi Ol O Gy.
 Source: epigenie.com
Source: epigenie.com
Designing Primers While good primer design is critical for successful PCR in any analysis designing primers for methylated DNA has many additional confounding factors to consider. Ad High precision and resolutionHigh efficiencyCost effective. You dont need to modify your sequence eg. OligoPerfect allows design up to 50 primers optimized for your reaction conditions. Hints For Optimizing Bisulfite Primer Design Zymo Guide.
 Source: sciencedirect.com
Source: sciencedirect.com
Use CpG island prediction for primer selection. Bisulfite Primer Seeker is a free program that has been optimized to design primers for CGrich sequences where others usually fail. Currently it can design primers for two types of bisulfite PCR. Multiple options for amplicon length and sequence location within the region of. A Bisulfite Free Approach For Base Resolution Analysis Of Genomic 5 Carboxylcytosine Sciencedirect.
 Source: geneious.com
Source: geneious.com
Features of the Bisulfite Primer Seeker Program. Unlike normal PCR bisulfite PCR primers need to be long usually between 26-30 bases and the amplicon size. CpG sites within the primers should be avoided or located at the 5-end with a mixed base at the cytosine position Y CT R GA. Ad High precision and resolutionHigh efficiencyCost effective. Primer Design Geneious.
 Source: researchgate.net
Source: researchgate.net
Bisulfite PCR amplification can be performed as a regular PCR reaction. Surprisingly no user-friendly primer design program is publicly available that could be used to design primers in plants and to simultaneously check the properties of primers such as the potential for primer-dimer formation. You can search various genomes with the designed primers to avoid non-specific PCR products by our fast ePCR method. Ad Design the perfect primers for your PCR CE sequencing and cloning. Evaluation Of The Targeted Bisulfite Pcr Sequencing Tbpseq Method Download Scientific Diagram.
 Source: semanticscholar.org
Source: semanticscholar.org
Designing Primers While good primer design is critical for successful PCR in any analysis designing primers for methylated DNA has many additional confounding factors to consider. The functions may be used sequentially or separately. BiSearch is a primer-design algorithm for DNA sequences. Multiple options for amplicon length and sequence location within the region of. Pdf Ultradeep Bisulfite Sequencing Analysis Of Dna Methylation Patterns In Multiple Gene Promoters By 454 Sequencing Semantic Scholar.
 Source: nature.com
Source: nature.com
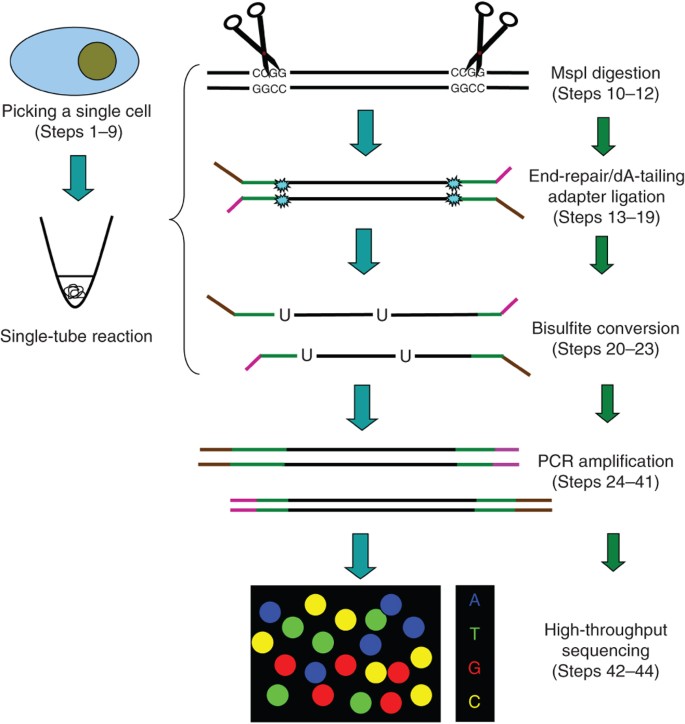
1 Methylation-Specific PCR MSP and 2 Bisulfite-Sequencing PCR BSP or Bisulfite-Restriction PCR. Use CpG island prediction for primer selection. B Primers for Bisulfite PCR are designed for subsequent sequencing and analysis of cytosines within the amplicon. OligoPerfect allows design up to 50 primers optimized for your reaction conditions. Profiling Dna Methylome Landscapes Of Mammalian Cells With Single Cell Reduced Representation Bisulfite Sequencing Nature Protocols.
 Source: researchgate.net
Source: researchgate.net
After bisulfite modification two DNA strands are no longer complementary. Although most of us have experience designing primers for traditional PCR designing effective primers for bisulfite-converted DNA poses challenges that must be overcome to perform this type of amplification successfully. This is based on the fact that DNA methylation is symmetric. Successful bisulfite primer design is critical to unbiased region-specific DNA methylation analysis. Flowchart Of Bisulfite Dna Sequencing Analysis Download Scientific Diagram.
 Source: sciencedirect.com
Source: sciencedirect.com
The software is able to design primers for the PCR amplification of both bisulfite-treated and untreated sequences. Primer design rules for MSP and bisulfite sequencing PCR. Ad High precision and resolutionHigh efficiencyCost effective. The sequences obtained from methylated gDNA and unmethylated gDNA are fundamentally different after bisulfite conversion. A Bisulfite Free Approach For Base Resolution Analysis Of Genomic 5 Carboxylcytosine Sciencedirect.
 Source: nature.com
Source: nature.com
CpG sites within the primers should be avoided or located at the 5-end with a mixed base at the cytosine position Y CT R GA. Use CpG island prediction for primer selection. 1 Methylation-Specific PCR MSP and 2 Bisulfite-Sequencing PCR BSP or Bisulfite-Restriction PCR. The software is able to design primers for the PCR amplification of both bisulfite-treated and untreated sequences. Targeted Bisulfite Sequencing Reveals Changes In Dna Methylation Associated With Nuclear Reprogramming Nature Biotechnology.
 Source: pinterest.com
Source: pinterest.com
The software is able to design primers for the PCR amplification of both bisulfite-treated and untreated sequences. General consideration for primer selection for bisulfite-conversion based PCR methods Because the two strands of DNA are no longer complementary after bisulfite modification strand-specific primers are used for PCR amplification. The detailed guidelines for primer design of bisulfite treated-DNA templates are discussed in Note 10. Designing primers for methylation PCRs. Epiquik Histone H3 Modification Multiplex Assay Kit Colorimetric Modification Kit Pattern.
 Source: nature.com
Source: nature.com
Existing primer design programs developed for standard PCR cannot handle primer design for bisulfite-conversion-based PCRs due to changes in DNA sequence context caused by bisulfite treatment and many special constraints both on the primers and the region to be amplified for such experiments. Ad Design the perfect primers for your PCR CE sequencing and cloning. This is especially recommended when primers are designed to amplify the highly redundant bisulfite treated sequences. How to cite MethPrimer. Library Free Methylation Sequencing With Bisulfite Padlock Probes Nature Methods.







